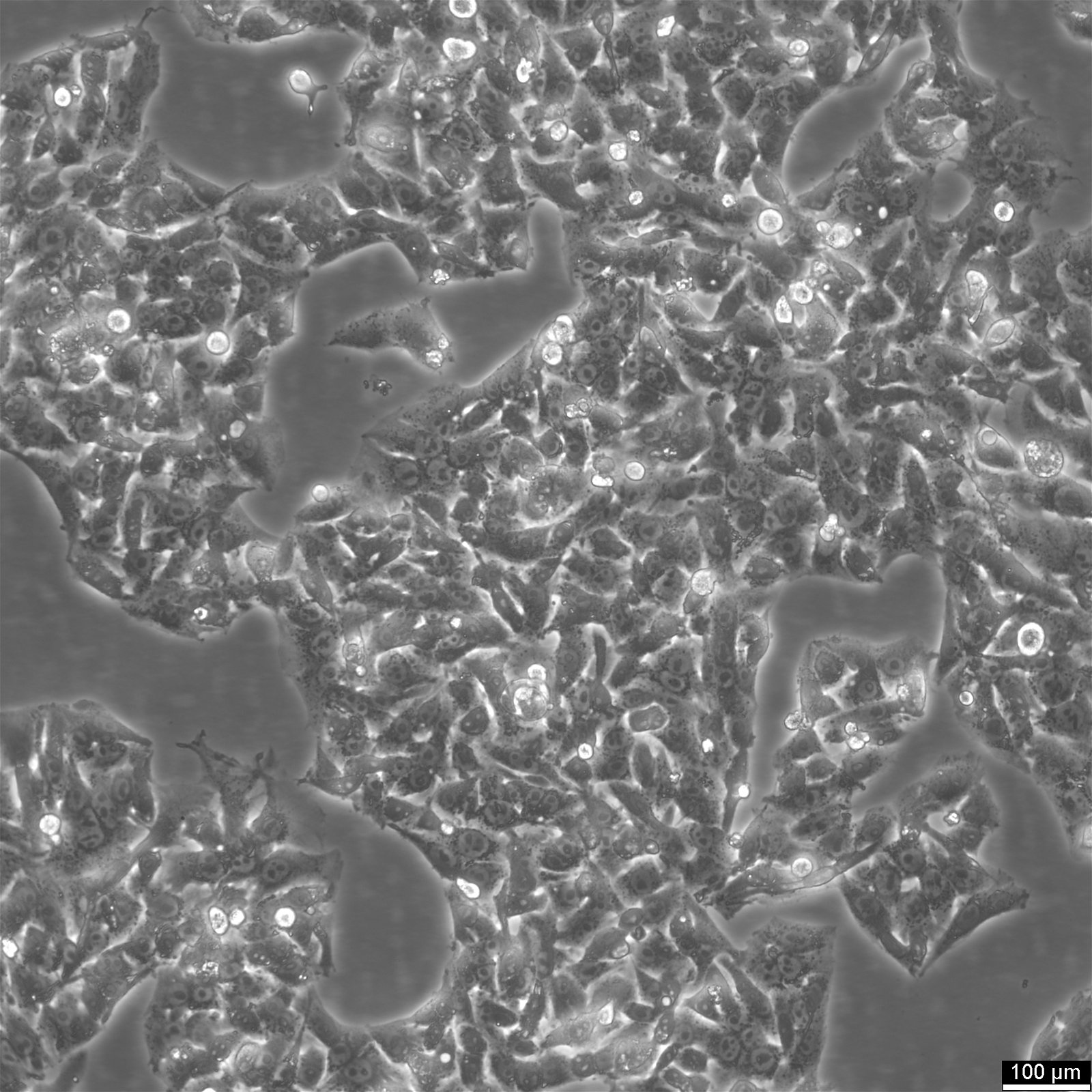
HepG2 Cells - A Liver Cancer Research Resource
Hep-G2 is a human liver cancer cell line originating from the liver tissue of a 15-year-old Caucasian male with hepatocellular carcinoma. These cells are frequently utilized in drug metabolism and hepatotoxicity studies. Although HepG2 cells have high proliferation rates and an epithelial-like appearance, they are non-tumorigenic and perform various differentiated hepatic functions. In 1975, researchers derived HepG2 cells from hepatocellular carcinoma, making it the first hepatic cell line to exhibit the critical characteristics of hepatocytes. In contrast to the previously established SK-Hep1 cell line, which lacks essential liver cell markers, HepG2 cells can secrete various plasma proteins and provide a valuable model for studying the intracellular dynamics of cell surface domains in human hepatocytes. These cells exhibit an epithelial-like morphology, have a modal chromosome number of 55, and can be stimulated with human growth hormone.
HepG2 Characteristics
Primary hepatocytes' typical shape is cubic and usually contains two nuclei. In contrast, HepG2 cells have an epithelial-like morphology with a single nucleus and a chromosome count ranging from 48 to 54 per cell. Although HepG2 cells can account for up to 25% of the total cellular protein, their size is larger than that of normal hepatocytes, making up about 10% of the complete protein in the cell. Cellular proteins are critical actors within the cell, executing the functions specified by genes.
Tumor cells, including those with an abnormal number of chromosomes, often exhibit an increase in the number of nuclei, up to seven per cell. Due to their high degree of differentiation in vitro, HepG2 cells provide an ideal model for studying the intracellular trafficking and dynamics of bile canalicular, sinusoidal membrane proteins, and lipids in human hepatocytes.
The average diameter of a HepG2 cell is around 10-20 µm, which is smaller than a hepatocyte with a diameter of 15 µm but similar to tumor cells with Hepatoblastoma (HB), which range from 10-20 µm.
HepG2 genetics
The Hep-G2 cell line exhibits several translocations, including those between the short arms of chromosomes 1 and 21, trisomies of chromosomes 2, 16, and 17, and tetrasomy of chromosome 20. The loss of the chromosome 4q3 region is also observed, associated with translocation t(1;4) often seen in Hepatoblastoma (HB) and other chromosomal abnormalities, such as trisomies 2 and 20. The number of chromosomes in HepG2 cells ranges from 50 to 60, indicating a hyperdiploid karyotype, while some cases exhibit more than 100 chromosomes and are characterized by tetraploid enlargement. HepG2 cells contain approximately 7.5 pg of DNA, 15% more than an average somatic cell. In comparison, primary hepatocytes have a cubic cell shape and typically contain two nuclei [1].
Mutational Profile of HepG2 Cells
The HepG2 cell line carries the TERT promoter region mutation C228T, also present in hepatocellular carcinoma (HCC) and hepatoblastoma (HB). This mutation contributes to immortalization by protecting telomeres in cancer cells. Additionally, HepG2 cells exhibit wild-type TP53, a critical gene for suppressing human cancer, as it plays a role in cell cycle arrest, apoptosis, and aging. Mutations in this gene can promote cell proliferation.
HepG2 cells participate in several pathways, including dysregulation of cell growth, survival pathways such as fetal and embryonal HB, and the Wnt/β-catenin pathway. Furthermore, the cell line has a characteristic deletion of the third exon of the CTNNB1 gene, which is identical to that seen in epithelial type HB [2,3].
Overview of HepG2 Hepatocellular Carcinoma Cells in Liver Research
HepG2 cells, originating from human hepatoma, have become an invaluable tool for researching liver functions and diseases, including hepatocellular carcinoma. These hepatic cell lines provide insights into the cellular responses of human hepatocytes under various experimental conditions. The use of luciferase reporter plasmids in HepG2 cells has been particularly effective for tracking gene expression and cellular transfections, which are fundamental in metabolic research, such as the study of ethanol's effects on liver cells.
Viral Infections and Liver Disease Studies Using HepG2 Cells
Immortalized hepatic tumor cell lines like HepG2 and Huh7 are essential in the study of viral infections, demonstrating complete cell cycle replication of hepatitis D (HDV) and expression of hepatitis B (HBV) [5,6]. In parallel, HepaRG cell lines play a critical role in elucidating HBV entry mechanisms [7]. HepG2 cells are also employed to investigate a variety of human liver diseases, from genetic conditions like progressive familial intrahepatic cholestasis (PFIC) and Dubin-Johnson Syndrome to environmental and dietary studies related to cytotoxic and genotoxic agents, as well as in drug targeting and hepatocarcinogenesis research [8,9]. Their use extends to trials with bio-artificial liver devices.
Interactions of HepG2 Cells with Biomaterials in Tissue Engineering
The interaction of HepG2 cells with various biomaterials is pivotal in tissue engineering. Techniques such as the colloidal probe technique help understand these interactions by measuring cell adhesion properties, which are vital in determining cell viability for the development of scaffolds and accurate liver tissue models.
Cell Behavior and Innovations in HepG2-Based Models
Studying cell behavior in HepG2-based models is crucial for liver disease research. Advancements in three-dimensional spheroid cell cultures have led to the creation of HepG2 cell spheroids, offering a more physiologically relevant model that closely mirrors normal hepatocytes. These 3D models, with increased metabolic activity, are indicative of the potential for HepG2 cells to serve as a model for hepatoblastoma and are significant in cancer treatment research, especially for simulating liver tumors and testing novel therapeutic approaches [10-12].
Comparison and Characteristics of HepG2 Among Other Tumor Cell Lines
HepG2 is one of the most widely used hepatic tumor cell lines, selected for its broad applications in scientific research among about 40 available hepatic tumor cell lines [13]. Despite its weak or absent expression of certain cytochrome P450 enzymes compared to normal hepatocytes, HepG2's metabolic profile has driven efforts to modify the cell line for better drug metabolism studies [13]. Compared to tumor cell lines like MCF7, PC3, 143B, and HEK293, HepG2 cells exhibit unique amino acid content profiles that significantly influence protein synthesis and secretion, highlighting their unique metabolic pathways [14].
Exploring Liver Disease Research with HepG2
Subculturing HepG2 Cells
Here are five steps for removing adherent cells from cell culture flasks using Accutase:
- Remove the medium from the cell culture flask and rinse the adherent cells using PBS without calcium and magnesium. Use 3-5 ml of PBS for T25 flasks and 5-10 ml for T75 flasks.
- Add Accutase to the cell culture flask, using 1-2 ml per T25 and 2.5 ml per T75 flask. Ensure that the Accutase covers the entire cell sheet.
- Incubate the flask at room temperature for 8-10 minutes.
- Carefully resuspend the cells with medium, using 10 ml of fresh medium.
- Centrifuge the resuspended cells for 5 minutes at 300xg, resuspend them in fresh medium, and dispense them into new flasks containing fresh medium.
Future Prospects for HepG2 Cells
The quest to unlock the full potential of the HepG2 cell line continues with groundbreaking progress in increasing the expression of cytochromes. Researchers are also exploring the possibility of three-dimensional spheroid cell cultures, which offer a more physiologically relevant system. The metabolic activity, including cytochromes, is remarkably higher in 3D spheroidal HepG2 models than in 2D cells, bringing us closer to creating a model that mirrors normal hepatocytes. Additionally, exploring the dynamic processes underlying the incorrect distribution of cell surface proteins can pave the way for a better understanding of liver diseases.
HepG2 Cells: Understanding Their Role and Distinctions in Biomedical Research - FAQs
References
- Vyas, R.C., Darroudi, F., Natarajan, A.T. Radiation-induced chromosomal breakage and rejoining in interphase-metaphase chromosomes of human lymphocytes, Mutat Res, 1991; 249(1):29-35.
- Woodfield, S.E., Shi, Y., Patel, R.H., Chen, Z., Shah, A.P., Srivastava, R.K., Whitlock, R.S., Ibarra, A.M., Larson, S.R., Sarabia, S.F., et al. MDM4 Inhibition: A Novel Therapeutic Strategy to Reactivate P53 in Hepatoblastoma. Sci. Rep. 2021, 11, 2967.
- Hussain, S.P., Schwank, J., Staib, F., Wang, X.W., Harris, C.C. TP53 Mutations and hepatocellular Carcinoma: Insights into the Etiology and Pathogenesis of Liver Cancer. Oncogene 2004.
- Schicht, G., Seidemann, L., Haensel, R., Seehofer, D., Damm, G. Critical Investigation of the Usability of Hepatoma Cell Lines HepG2 and Huh7 as Models for the Metabolic Representation of Resectable Hepatocellular Carcinoma. Cancers 2022, 14(17), 4227.
- Verrier, E.R., Colpitts, C.C., Schuster, C., Zeisel, M.B., Baumert, T.F. Cell Culture Models for the Investigation of Hepatitis B and D Virus Infection. Viruses 2016, 8, 261.
- Verrier, E.R., Colpitts, C.C., Bach, C., Heydmann, L., Weiss, A., Renaud, M., Durand, S.C., Habersetzer, F., Durantel, D., AbouJaoudé, G., et al. A Targeted Functional RNA Interference screen Uncovers Glypican 5 as an Entry Factor for Hepatitis B and D Viruses. Hepatology 2016, 63, 35–48.
- Gripon, P., Rumin, S., Urban, S., Le Seyec, J., Glaise, D., Cannie, I., Guyomard, C., Lucas, J., Trepo, C., Guguen-Guillouzo, C. Infection of a Human Hepatoma Cell Line by Hepatitis B Virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660.
- Mersch-Sundermann, V., Knasmüller, S., Wu, X.J., Darroudi, F., Kassie, F. Use of a human-derived liver cell line for the detection of cytoprotective, antigenotoxic and cogenotoxic agents. Toxicology. 2004; 198(1–3): 329–340.
- Fanelli, A. HepG2 (liver hepatocellular carcinoma): cell culture. HepG2. Retrieved 3 December 2017.
- Xuan, J., Chen, S., Ning, B., Tolleson, W.H., Guo, L. Development of HepG2-Derived Cells Expressing Cytochrome P450s for Assessing Metabolism-Associated Drug-Induced Liver Toxicity. Physiol. Behav. 2017, 176, 139–148.
- Ooka, M., Lynch, C., Xia, M. Application of in Vitro Metabolism Activation in High-Throughput Screening. Int. J. Mol. Sci. 2020, 21, 8182.
- Huang, L., Coughtrie, M.W.H., Hsu, H. Down-Regulation of Dehydroepiandrosterone Sulfotransferase Gene in Human Hepatocellular Carcinoma. Mol. Cell. Endocrinol.
- Zhu, Z., Hao, X., Yan, M., et al. Cancer stem/progenitor cells are highly enriched in CD133 + CD44 + population in hepatocellular carcinoma. Int J Cancer. 2010; 126:2067-2078.
- Arbus, C., Benyamina, A., Llorca, P.-M., Baylé, F., Bromet, N., Massiere, F., Garay, R.P., Hameg, A. Characterization of human cytochrome P450 enzymes involved in the metabolism of cyamemazine. Eur J Pharm Sci. 2007 Dec;32(4-5):357-66.